|
Ghanshyam is an AI Research Scientist at Intel Labs in Bangalore, India. His work focuses on AI for Genomics, High-Performance Computing, and Computational Biology. Explore my Image Gallery My Erdős number is 5 Email / CV / LinkedIn / Twitter / Github / Google Scholar / Blog |

|
| [January 2026] Our paper titled "Accelerating minimap2 for whole-genome alignment" is accepted at Oxford Bioinformatics. |
| [December 2025] Our paper is featured on the cover of Genome Research. |
| [November 2025] Released DipGenie v1.0. Our work titled “Haplotype-resolved diploid genome inference on pangenome graphs” is now online. |
| [July 2025] Our paper, titled "Pangenome-based genome inference using integer programming" has been accepted for Genome Research’s RECOMB special issue. |
| [June 2025] Released mm2-plus v1.2 based on Minimap2-2.30 (r1287) code-base. |
| [May 2025] Released mm2-plus v1.1 based on Minimap2-2.29 (r1283) code-base. |
| [April 2025] I will be giving talk at RECOMB 2025, held at Seoul, South Korea. |
| [January 2025] PhD Colloquium done! Joined [Intel Labs] as AI Research Scientist. |
| [December 2024] Our paper titled "Integer programming framework for pangenome-based genome inference" is accepted at RECOMB 2025 (acceptance rate 16%). Had a fireside chat with Dr. Heng Li (HMS & DFCI) discussing the future of pangenomics. [Image] |
| [November 2024] Preprint on "Accelerating whole-genome alignment in the age of complete genome assemblies". [preprint] [code] |
| [October 2024] Preprint on "Integer programming framework for pangenome-based genome inference". [preprint] [code] |
| [August 2024] Our research "New algorithms advance genomic equity and personalized medicine" is highlighted by Indian Institute of Science and The Times of India. |
| [July 2024] Our work "Haplotype-aware sequence alignment to pangenome graphs" is now online at Genome Research. |
| [May 2024] Serving as a PC member for AccMLBio (ICML 2024). |
| [April 2024] I will be giving talks at RECOMB-Seq 2024 and RECOMB 2024, both held at MIT, USA. |
| [March 2024] Our work on "Accelerating whole-genome alignment using parallel chaining algorithm" is accepted for short talk at RECOMB-Seq 2024. |
| [February 2024] Invited talk by IEEE on "Why Use Human Genome Graphs as a Reference? Insights into Scalable Genome Graph Algorithms." [Poster] [Memento] |
| [December 2023] Our paper titled "Haplotype-aware Sequence-to-Graph Alignment" is accepted at RECOMB 2024. [Link] |
| [November 2023] Preprint on "Haplotype-aware Sequence-to-Graph Alignment". [preprint] Minichain (v1.3) released with support for "Haplotype-aware Sequence-to-Graph Alignment". [Link] |
| [August 2023] Received Intel Research Fellowship 2023-24. |
| [June 2023] Our paper titled "Co-linear Chaining on General Graphs" is accepted at WABI 2023. |
| [May 2023] Released, PanAligner: Long read mapper to a pangenome graph. Link. |
| [April 2023] I gave a talk on Sequence to Graph Alignment using Gap-Sensitive Co-Linear Chaining at RECOMB 2023. I presented a poster titled "Minichain: A New Method for Pangenome Graph Construction" at RECOMB-SEQ 2023. I presented my research work on Pangenomes at the EECS 2023 Symposium. |
| [March 2023] Received RECOMB 2023 Travel Fellowship. Our poster titled "Minichain: A new method for pangenome graph construction" has been accepted at RECOMB-SEQ 2023. [Link] |
| [Feb 2023] Minichain v1.2 released. [Link] Minichain v1.1 released. [Link] |
| [Dec 2022] Presented a poster titled "Scaling sequence-to-DAG alignment with parameterized gap-sensitive co-linear chaining algorithms" at HiPC 2022. [Link] |
| [Nov 2022] A paper titled "Sequence to graph alignment using gap-sensitive co-linear chaining" is accepted at RECOMB 2023. [Link] |
| [Dec 2021] Winner: National HPC Hackthon 2021. [Link] |
| [Oct 2021] Benchmarking human whole genome and RNA sequencing using Nvidia Ampere A100 GPUs. [Link] |
| Minichain: Long-read aligner to pangenome graphs. |
| PanAligner: Long read aligner for cyclic and acyclic pangenome graphs. |
| PHI: Pangenome graph-based genome inference. |
| mm2-plus: Fast long-read and whole-genome aligner. |
| DipGenie: Haplotype resolved diploid genome inference using pangenome graphs. |
|
|
 |
Haplotype-resolved diploid genome inference on pangenome graphs. [preprint] [code] Ghanshyam Chandra, William T. Doan and Daniel Gibney. |
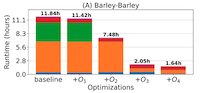 |
Accelerating minimap2 for whole-genome alignment. [preprint] [code] Ghanshyam Chandra, Md. Vasimuddin, Sanchit Misra and Chirag Jain, (To be published in Oxford Bioinformatics) |
 |
Integer programming framework for pangenome-based genome inference. [preprint] [code] Ghanshyam Chandra, Md Helal Hossen, Stephan Scholz, Alexander T Dilthey, Daniel Gibney and Chirag Jain, (Appeared in RECOMB 2025, Published in Genome Research 2025) |
 |
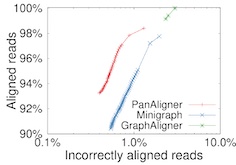
Haplotype-aware sequence alignment to pangenome graphs. [paper] [code] Ghanshyam Chandra, Daniel Gibney and Chirag Jain, (Appeared in RECOMB 2024, Published in Genome Research 2024) |
 |
Co-linear Chaining on Pangenome Graphs [paper] [code] Jyotshna Rajput, Ghanshyam Chandra and Chirag Jain, (Appeared in WABI 2023, Published in Algorithms for Molecular Biology) |
 |
Gap-Sensitive Colinear Chaining Algorithms for Acyclic Pangenome Graphs. [paper] [code] Ghanshyam Chandra and Chirag Jain, (Appeared in RECOMB 2023, Published in Journal of Computational Biology) |
|
Cloned from Jon Barron. |